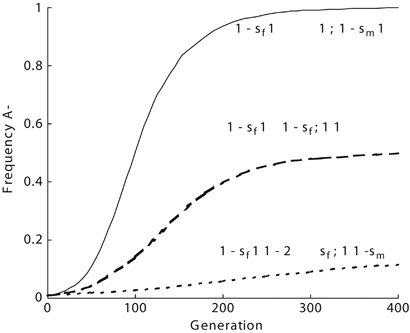
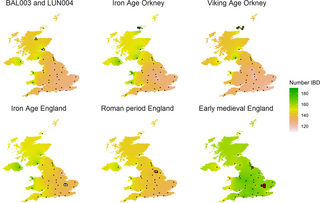
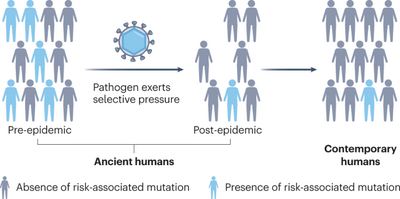
“Overall, the original ‘malaria hypothesis’ of Haldane that diseases such as thalassemia are polymorphisms with an advantage to heterozygotes in malarial environments has been proven correct.” #dna #isogg #malaria #epigenetics #population #genetics #geneadons